TransBTS: Multimodal Brain Tumor Segmentation Using Transformer
TransBTS
During the preparation phase of my graduation project, I was surveying a lot of research papers around Brain Tumor Segmentation, and I found this paper; TransBTS, which uses transformers in MRI(magnetic resonance imaging) scans segmentation. Commonly, transformers have been widely used to solve Natural Language Processing tasks, however it has been used with a convolutional neural network to solve this segmentation task.
1-Glioma:-
The most common brain tumor which consists of 67% of the brain tumor types.
In the USA, around 24,000 are caught up with glioma; 10,000 are females and 14,000 are males.
Doctors are preferably into dividing the tumor into 4 parts:-
1-Background(brain)
2-Whole Tumor
3-Enhanced Tumor
4-Tumor Core
2-Introductions to transformers:-
Recurrent neural networks(RNNs) have been used to solve NLP problems which takes input string of a phrase and starts putting them in the network to feed it word by word by forward and backward propagation to update weights and reduce the loss function, however it always ends up with the vanishing gradients problem and fixed it by using LSTM(long short term memory) and GRUs(gated recurrent unit) to pick which long/short memory will be kept.
The following figure illustrates the structure of RNN, LSTM and GRU.
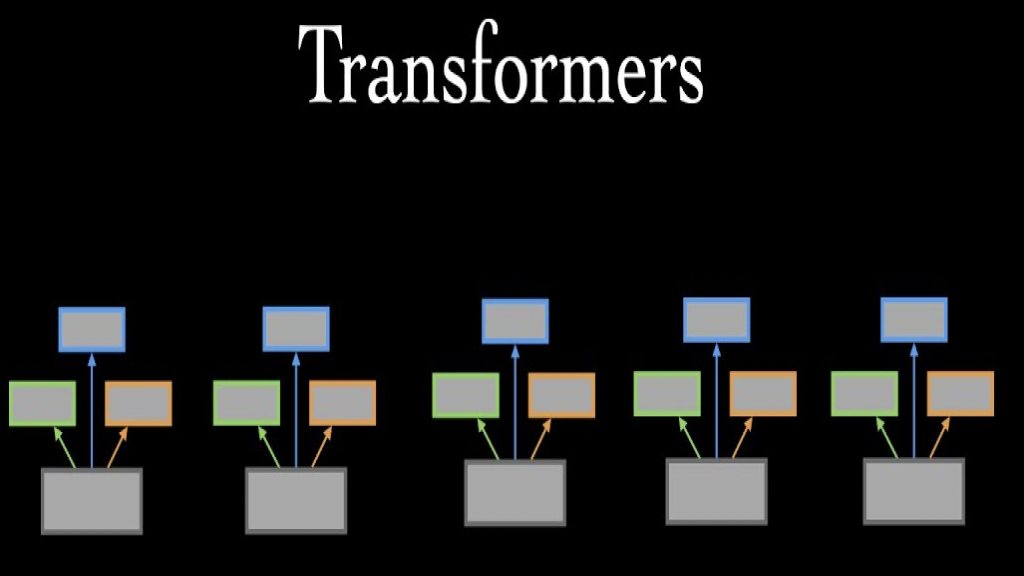
Transformers adopt the mechanism of self-attention differentially weighting the signifi- cance of each part of the input data.Transformers are used primarily in natural language processing as they handle sequential data like Recurrent Neural Networks (RNN) but unlike RNNs that process data in-order from their name “Recurrently”, Transformer’s attention mechanism provides context for any position in the input sequence. This helps with better parallelization of jobs and reducing training time. Also, Transformers capture global features better as the information does not vanish like in RNNs due to processing data recurrently.
3-Why use visual transformers?
The problem with the adoption of Transformers in computer vision is the complexity of computations that is in order of O(N2) because the self-attention is getting context information for each word with respect to all the words in the sequence so each token is one pixel so for a 100x100 grayscale image this is 108 computations which is huge number of computations for a small image. Visual Transformers approach this problem by dividing the image to tokens and these tokens are 16x16 in size this reduces the number of computations from 108 to 1600. The usage of these small tokens can capture global context in the image.
4-UNet3D:-
The architecture of UNet is mainly composed of 3 things; encoder path, bottleneck and decoder path.
In the encoder path the input data is compacted and the local features are learned throughout the process using convolutional kernels until it reaches the bottleneck, then during the decoder path, the network uses reverse convolution to learn the deep features and concatenates with the information with the similar level in the encoder path as shown in the figure below.
5-TransBTS architecture:-
The model here extracts both the benefits of UNet encoder to compact the information to be right fed to the visual transformer as shown:
After passing the information, glioma is then segmented in a set of slices which is then reformed in the shape of MRI and displayed, the scaling happens during the encoder path, reshaped, and then embedded to be fed right to the transformer which uses MHA(multi head attention) and feed forward network.
The equations used in transformers are the following:-
The equation introduced the learnable PE and added them to the feature map f for features embeddings where W is the linear projection operation, and for each layer;
MHA denotes multi-head attention of the transformer, as LN denotes layer normalization.
Finally the feed forward(FFN) of the normalized layer(LN) is calculated to the final output of feature embedding of the transformer and then passed to the decoder path.
6-Model Summary:-
7-Results:-
The datasets used in the experiment are BraTS 2019 and BraTS 2020, for the BraTS 2019 validation set their proposed model achieved the following metrics: Dice ET=78.93, WT=90.00 TC=81.94, HD ET=3.736, WT=5.644, TC=6.049. FOr the BraTS 2020 validation set, they achieved: Dice ET=78.73, WT=90.09, TC=81.73, HD ET=17.947, WT=4.964, TC=9.769 The authors addressed the challenge of computations cost as the transformers use a lot of computa- tions.
Summary:-
The problem with the MRI segmentation task of the glioma tumor.
Visual transformers and Unet fusion in the TransBTS network.
Used architecture and summary of the model.
Results after training on BraTS challenge dataset.
- Youssef Khaled
- Mar, 25 2022